Zavodszky lab
Research
Ligand-induced changes in proteins
Ligand binding induces changes in the receptor protein, providing a way to regulate protein function. Ligand-induced changes can be either localized to the vicinity of the binding site, or can be long-range, as in the case of allosteric proteins. In this latter class of proteins, binding of a ligand to a regulatory site alters the activity at a distant active site with some kind of a signal believed to be transmitted from one site to the other. Experimentalists are trying to pinpoint the residues transmitting such signals by systematically mutating amino acids between the two sites, while theoreticians are looking for channels or pathways that would allow some kind of mechanistic communication between the two sites.
An alternative idea is that changes in flexibility and dynamics upon ligand binding are responsible for the observed effects. Network representations of biomolecules, as well as normal mode analysis and molecular dynamics simulations can be used to study the flexibility and dynamics of these systems.
Poster presented at the Steenbock symposium (May 2006) on this topic
Flexibility at protein-protein interfaces
The ability to reconstruct and study biological pathways greatly depends on how well we can predict the mode of interaction between the elements of the pathways, which are usually proteins of various functions. Given the structures of the free proteins, current computational docking tools cannot predict the structure of the bound complex when one or both or the binding partners undergo large conformational changes. Based on flexibility analysis of the unbound structure using the graph-theoretical algorithm FIRST, it is possible to predict what parts of the protein are most likely to undergo conformational changes. Large-scale movements of the predicted flexible parts can be modeled using clever algorithms that sample only the low frequency modes of the possible motions.
Structure-based computational screening
We are working on identifying new ligands/inhibitors by computational screening to a number of different protein targets relevant for disease therapy:
- Urokinase-type plasminogen activator (uPA) plays a key role in tumor invasion and metastasis. High levels
of uPA in primary tumor tissues are associated with unfavorable prognosis with high risk of disease
recurrence.
- Arachidonic acid metabolizing lipoxygenase (P-12-LOX) is involved in prostate cancer progression: the level
of P-12-LOX expression increases at advanced stages of the disease; overexpression of P-12-LOX in
human prostate cancer cells stimulates angiogenesis and tumor growth; an inhibitor of P-12-LOX was found
to slow down metastasis.
- RbgA is an essential GTPase in the assembly of ribosomal subunit 50S in the bacterium Bacillus subtilis.
Small molecules or peptides that bind to this protein can disrupt ribosome biogenesis. This is a project in
collaboration with Rob Britton from the Department of Microbiology at MSU (http://www.msu.edu/~rbritton/).
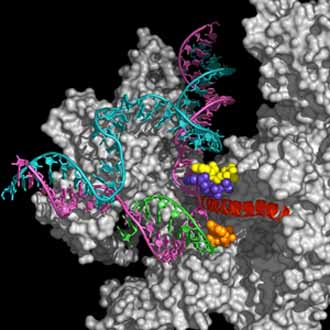
Flexibility and dynamics of eukaryotic RNA polymerase II
|
RNA polymerase II is responsable for transcribing the genetic material encoded in the genes into mRNA, which, on its turn, provides the blueprint for protein synthesis. The multisubunit RNA polymerase II is a large and complex machine with multiple functions: grabs the double stranded DNA, slides along it until finding the promoter. It separates the two DNA strands creating a "transcription bubble", binds single ribonucleotides complementary to the single stranded DNA template, and connects them by phosphodiester bonds to form the RNA chain. When it reaches the termination site, releases the newly formed RNA molecule. While it is doing all this, it has to recognize and interact with various transcription factors that regulate this process. A complex methodology is designed combining MD simulation, flexibility and normal mode analysis to model the motion of this complex system.
Poster presented at the Indy07 symposium (May 2007) on this topic
|
|
Research in college science teaching: Developing a Biochemistry Concept Inventory
What kind of understanding do undergraduate students possess about the biochemical processes underlying life? What is the impact of the undergraduate biochemistry courses on their perceptions? What kind of problems do they struggle with as they try to cope with the ever increasing amount of information finding its way into general biochemistry teaching materials? The objective of my work is to develop an assessment tool in form of a Biochemistry Concept Inventory (BCCI) that helps answering these questions.
The starting point of a concept inventory is the assembly of a list of key concepts and organizing principles of a topic area, followed by the development of a multiple-choice test to assess conceptual understanding of students of these ideas. The list of key concepts from the BCCI will reflect a broad consensus among expert opinions in the field. In collaboration with education researchers affiliated with CRCSTL and educators from the department of Biochemistry and Molecular Biology, I will identify student misconceptions through qualitative and quantitative analysis of student interviews and answers to open-ended questions related to the key concepts. The BCCI will be designed to test for conceptual understanding instead of recall of vocabulary or isolated bits of information. For the purpose of this work, we define conceptual understanding as developing an integrated and functional grasp of concepts. The topics of the questions of the BCCI will be focused on the key concepts identified previously. Distractors will be selected from student responses written to open-ended questions and answers given to questions asked during interviews that reveal misconceptions, alternative conceptions or lack of foundation in basic chemistry or biology.
The BCCI will be used to identify and compare problems that hinder student understanding of biochemistry at three different levels of general biochemistry courses at Michigan State University.
This work has been started under the guidance of the Center for Research in College Science Teaching and Learning (CRCSTL) at MSU.